THE POPULARITY OF NMR
There is no doubt that NMR spectroscopy is currently the most popular technique for characterizing various types of samples in general and particularly in unraveling molecular structures for the following reasons:
- NMR is applicable to liquid, solid or semi-solid samples on the one and to small, medium and large molecules on the other side
- Almost any element in the periodic table may act as a nuclear sensor and in particular the various interactions between nuclei may be detected and measured both qualitatively and quantitatively not only for purified single component samples but also for mixtures such as biological extracts. Hence NMR has become the go-to technique for scientists looking for an information-rich and non-destructive anayltical tool to reveal the structure, identiy, concentration and behavior of such kind of samples
- Particularly for high resolution NMR spectroscopy – the subject of this tutorial – numerous pulse experiments have been designed (“tailored” pulse sequences) for any type of liquid sample to exploit these nuclear properties. As a result the structural information – describing not only the “static” shape but also and in particular the “dynamic” behavior of molecules or reaction processes – now available with NMR spectroscopy is probably greater and more readily obtainable than with any other single technique
- Tremendous technical improvements in NMR spectrometer design over the last few years, with the availability of increasing magnetic field strengths, ultrashielded superconducting magnets, more sensitive and more convenient probe-heads and spectrometers characterized by highest stability and precision as the most prominent have improved steadily the quality of the spectral parameters, their reproducibility and thus the reliability of the structural information derived from it
- A rapid increase in computer power to enable software to be developed that improves spectrometer control and simplifies data processing which opens up NMR also to non-experts
"MAGNET STORE"  MODERN NMR SPECTROMETER (BRUKER)  |
- Of course, several other methods for investigating molecular structures exist such as the well-established and successful x-ray crystallography or the very recent highly promising cryo-electron microscopy (cryo-EM), which allows individual molecules to be “directly observed”. However, none of these techniques yields that detailed information on a molecule with respect to its atomic network, its stereochemistry, its conformation and its dynamics to solve “simple” daily problems, such as e.g. a cis/trans-clarification on the one side and to establish the structure of a protein on the other side.
Furthermore, the experimental time and effort with this technique is moderate, measuring times – at least for routine investigations – are reasonable despite the technique’s inherent low sensitivity. Subsequent data processing is not that demanding, the complexity of final analysis and interpretation is typically straightforward.
These attributes are in an excellent proportion to the manifold of valuable structural information available with NMR and explains its high attractiveness and its large number of users.
A NMR TUTORIAL - WHY ?
The occasion and motivation for providing this NMR-Tutorial is as follows:
- Undoubtedly and in order to best exploit the potential of modern NMR spectrometers for elucidating molecular structures the user must be well versed in the precise interpretation of spectral parameters extracted from the final NMR spectrum. However, NMR spectra and hence the spectral parameters derived thereof depend not solely on the structure of the investigated molecule(s) but in particular also on the NMR experiment and the mode of data processing applied and the corresponding experimental and processing parameters, a fact that is often neglected, if at all addressed and dealt with.
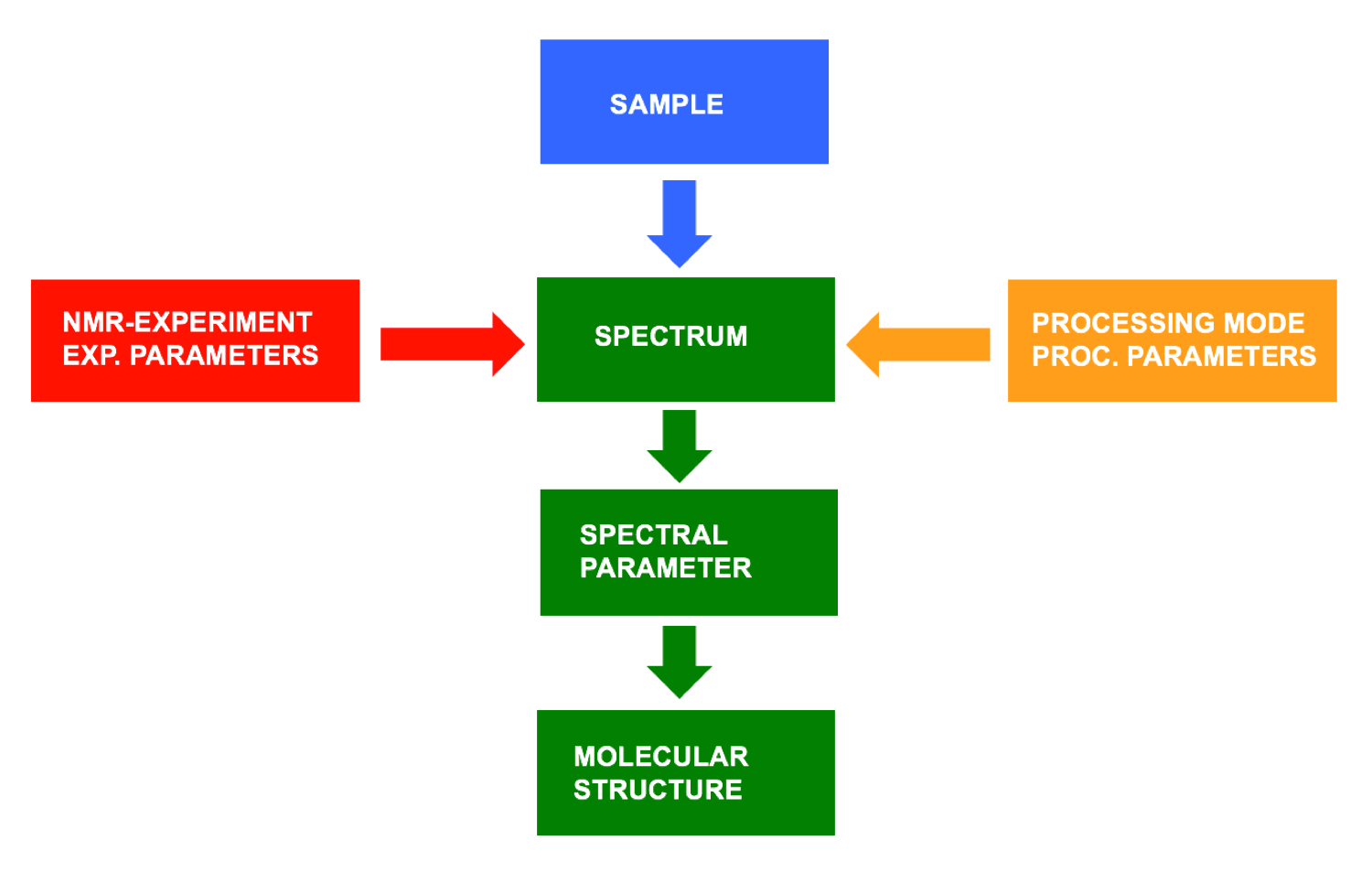 |
Therefore, especially with more demanding NMR investigations and when applying more sophisticated NMR pulse sequences the user should have the necessary expertise to fully understand how the corresponding NMR experiments work, how their outcome depends on the experiment’s parameters and how the different types of spectra arise. In addition – to keep this always in mind – the values and quality in general of NMR spectral parameters such as signal intensities or coupling constants depend also on the kind and the corresponding parameters of data processing. Consequently, although this is not the subject of this tutorial, adequate data processing is an important prerequisite for reliable structure elucidation. In textbooks, instructions and recipes for the correct analysis and interpretation of NMR spectra are often associated with the motto "to make the most of your NMR data". Instead, and since the selection of the NMR experiments and the correct choice of the corresponding measurement parameters must be included, the motto should be (re)formulated more comprehensively as: "To make the most of your sample and the available NMR spectrometer time". This is exactly the aim of this tutorial. |
- The benefits of automation in NMR spectroscopy such as standardization, sample throughput or convenience are beyond dispute but there are also a few disadvantages. With automated NMR spectrometers the course of an experiment, its setup and its parameter dependence are covered in some sense by a “black box” making the corresponding expertise at a first glance apparently dispensable, which however is not true (see before). Indeed and unfortunately there is evidence that within the NMR spectroscopist‘s community the necessary theoretical background and skills for thoroughly understanding the mechanics of pulse sequences or the relation between experimental parameters such as repetition rates or pulse angles and the spectral outcome more and more get lost. Consequently, there is concern that without this expertise not only the “beauties” of modern NMR experiments may be overlooked but more severe the necessary care and critical validation of experimental results, an indispensable prerequisite for reliable structure elucidation, will be missing. One should take these concerns seriously particularly in view of the emergence and the potential of the so-called artificial intelligence (AI). AI will most likely revolutionize NMR spectroscopy in general, will not be restricted to spectra-analysis and -interpretation but will certainly include also the choice of experiments, their set-up and the spectrometer control.
- With the versatile BRUKER TOPSPIN software package powerful and convenient tools for NMR data acquisition, processing and analysis are available with a free academia (processing only) version. Among the various modules NMRSIM – a virtual NMR spectrometer (or “NMR-flight-simulator”) dedicated for the numerical simulation of 1D and 2D NMR experiments – is likely the most attractive and most promising component of this NMR tutorial. In combination with TOPSPIN’s processing and analysis modules the simulated NMR raw data may be efficiently processed, inspected and evaluated. This provides the highly valuable basis for teaching the user the necessary skills, particularly without making demands on valuable measuring time on a real NMR spectrometer.
- Combined with the corresponding “script” (the Tutorial Guide) in hand the Tutorial will hopefully pique the user’s curiosity and will convince him to not be satisfied solely with the fact t h a t a NMR experiment works, but to wonder w h y and h o w it works.
THE TUTORIAL'S AUDIENCE AND THE USER'S PREREQUISITES
The NMR tutorial is devised as an autodidactic tool for adopting or extending theoretical and practical skills in NMR spectroscopy and for building up the corresponding competences for operating a NMR spectrometer.
Although laid-out for NMR newcomers the tutorial assumes that the user is familiar with the basics of NMR as given in introductory lectures on NMR spectroscopy or in corresponding textbooks. On the other hand, the tutorial is also designed to be an advanced training tool for users having already gained first experiences with spectrometer operation, i.e. for users wanting to engross their comprehension for the relation between theory and practice or the dependence of the spectral appearance on experimental parameters.
Therefore, choose and concentrate on those issues, which are of interest for you, which may best fulfill your needs and feel free to skip those parts you are already familiar with.
Although laid-out for NMR newcomers the tutorial assumes that the user is familiar with the basics of NMR as given in introductory lectures on NMR spectroscopy or in corresponding textbooks. On the other hand, the tutorial is also designed to be an advanced training tool for users having already gained first experiences with spectrometer operation, i.e. for users wanting to engross their comprehension for the relation between theory and practice or the dependence of the spectral appearance on experimental parameters.
Therefore, choose and concentrate on those issues, which are of interest for you, which may best fulfill your needs and feel free to skip those parts you are already familiar with.
THE TUTORIAL'S GOALS
The main goals pursued with this tutorial may be summarized as follows:
- Concentrating on data acquisition and emphasizing practice build up your NMR competence in general, perfect your skills for operating a NMR spectrometer, for processing the acquired data and for a critical assessment of the experiments outcome and the spectroscopic results.
- Improve your understanding of the “mechanics” of 1D and 2D NMR experiments on a theoretical and practical level in detail
- Understand the effects and the mutual relationship of pulse sequence elements such as rf-pulses or magnetic field gradients and corresponding parameters on the one and chemical shifts, coupling and relaxation on the other side and their influence on the final “spin’s response”.
- Experience the potential, scope and the limits of NMRSIM – the virtual NMR spectrometer – for autodidactic education, (own) teaching and particularly for your research. Let you stimulate to experiment yourself, to perform corresponding simulative studies for investigating other standard pulse sequences, their parameter dependence, for modifying pulse sequences or for designing and testing your own 1D or 2D experiments.
- Let your curiosity and interest in NMR experimentation be piqued and lay a solid basis for this powerful analytical tool.
- Most important marvel at the beauties of the NMR physics. “Enjoy” the fundamentals of this fascinating technique instead of just "learning" it !
THE TUTORIAL'S TOPICS
The Tutorial Guide consists of four parts:
- ABOUT THIS TUTORIAL: Gives a survey about the tutorial's elements and scope and the themes of the individual chapters and specifies the typical issues of the main chapters (dedicated to a representative NMR experiment). The sequential steps of a typical session (dedicated to a selected experiment/simulation) as part of such a main chapter and the interaction with the TOPSPIN software and the verification data is outlined. Furthermore, an overview about a few format elements making the tutorial's handling more convenient and the recommendation to keep a laboratory journal, are given.
- THE BASICS OF NMR IN BRIEF: Deals in brief with a few more general issues such as the aims of NMR spectroscopy, the dependence of NMR spectral parameters on molecular features, the classification of NMR experiments, the 1D- / 2D-concept and the analysis/simulation of NMR pulse sequences. This part outlines also the common strategy and the classical steps of a NMR investigation and addresses the concept and main elements of 1D and 2D NMR data processing. Hence this chapter of the tutorial guide aims for basic “refreshing” and serves to stimulate the user for the subsequent, highly interactive main part of the tutorial dedicated to a set of representative 1D and 2D NMR experiments.
- SOFTWARE MODULES: Addresses the download and installation of the BRUKER TOPSPIN software package and gives short interactive introductions to its processing/analysis and simulation (NMRSIM) modules. Users who are not familiar with the simulation of NMR experiments and data processing are guided on a short tour through the individual steps for simulating 1D and 2D experiments and for processing / analyzing the corresponding simulation data. This allows the newcomer to gain his first hands-on experience with these tools and to build up personal skills for reliable and efficient NMR simulation and NMR data processing. Note at this stage that NMRSIM yields NMR raw data (time domain signal) and that quite obviously the outcome of simulations can only be interpreted on the basis of NMR spectra (frequency domain signal). Therefore, adequate NMR data processing is indispensable and will be part of this tutorial. Furthermore, instructions for the download and storage of simulation files that will be used in the further course are given.
- REPRESENTATIVE 1D AND 2D NMR EXPERIMENTS: In this main part of the tutorial the user becomes acquainted with a few 1D and 2D pulse sequences and – by taking advantage of guided simulations – experiences the functioning and the parameter dependence of these experiments. The simulations concentrate on the one side on more general, experiment comprehensive topics and on the other side on experiment’s specific issues such as its set-up and its specific characteristics. This part is highly interactive taking advantage of so-called “Check its”, allowing the user to experience a given experiment in detail, to evaluate its response to individual parameter settings and to study and interpret the spectral outcome. For comparison and for verifying its own conclusions and competence reference NMR spectra and reference findings for the individual “Check-its” are available.
THE TUTORIAL'S LAYOUT
The tutorial appears in an electronic form and is accessible via the central teaching and learning platform ILIAS administrated by the Support Center for ICT-Aided Teaching and Research of the University of Bern (iLUB).
To become familiar with the theoretical and practical aspects of NMR spectroscopy the tutorial is designed as a highly interactive online-learning platform.
Due to this interactive approach suitable NMR software tools (BRUKER TOPSPIN package) for simulation and data processing - together with corresponding parameter- and reference data-files for verification - will be installed and operated locally. Hence, the use of a computer with adequate capacities, i.e. a PC (Windows / Linux) or MAC (macOS) is not only desirable but mandatory.
Taking advantage of BRUKER software tools and BRUKER pulse sequences the tutorial is inevitably manufacturer-oriented. Nevertheless its layout is hold as general as possible keeping the tutorial open for interested users working with spectrometers of different origin.
To become familiar with the theoretical and practical aspects of NMR spectroscopy the tutorial is designed as a highly interactive online-learning platform.
Due to this interactive approach suitable NMR software tools (BRUKER TOPSPIN package) for simulation and data processing - together with corresponding parameter- and reference data-files for verification - will be installed and operated locally. Hence, the use of a computer with adequate capacities, i.e. a PC (Windows / Linux) or MAC (macOS) is not only desirable but mandatory.
Taking advantage of BRUKER software tools and BRUKER pulse sequences the tutorial is inevitably manufacturer-oriented. Nevertheless its layout is hold as general as possible keeping the tutorial open for interested users working with spectrometers of different origin.
ACKNOWLEDGEMENTS
I would like to express my gratitude to BRUKER BioSpin for their interest in this project and corresponding support.
I am indebted to Dr. T. Thiel and Dr. M. Engelhardt for providing active and stimulant initial assistance, Dr. P. Kessler and Dr. B. Guigas - responsible for the TOPSPIN software modules - for their helpful advices and excellent collaboration.
For the project’s implementation into an electronic form and its finalization I am very grateful to Y. Seiler and H. Lauener (ILUB University of Berne) for their most cooperative and competent support.
I am indebted to Dr. T. Thiel and Dr. M. Engelhardt for providing active and stimulant initial assistance, Dr. P. Kessler and Dr. B. Guigas - responsible for the TOPSPIN software modules - for their helpful advices and excellent collaboration.
For the project’s implementation into an electronic form and its finalization I am very grateful to Y. Seiler and H. Lauener (ILUB University of Berne) for their most cooperative and competent support.
